SCAMP Profile Types
SCAMP can compute various types of matrix profiles. Each has slightly different semantics and provide different ‘views’ of the distance matrix. All of these profile types support AB-joins.
- Nearest Neighbor and Index:
This is the default profile type and the normal definition of matrix profile, it will produce the nearest neighbor distance/correlation of every subsequence as well as the index of the nearest neighbor
CLI flag:
--profile_type=1NN_INDEXpyscamp functions: selfjoin, abjoin
- Nearest Neighbor Only:
This is a slightly faster version of the default profile type, but it only returns the nearest neighbor distance/correlation not the index of the nearest neighbor
CLI flag:
--profile_type=1NNpyscamp functions: Currently unsupported.
- Sum of correlations above threshold [correlation histogram]:
Rather than finding the nearest neighbor, this profile type will compute the sum of the correlations above the specified threshold (
--threshold) for each subsequence. This is like a frequency histogram of correlations.CLI flag:
--profile_type=SUM_THRESHpyscamp functions: selfjoin_sum, abjoin_sum
- Approximate K nearest neighbors:
[EXPERIMENTAL, GPU ONLY, DISTRIBUTED UNSUPPORTED] This returns the approximate K (
--max_matches_per_column) nearest neighbors and their correlations/indexes for each subsequence. A threshold (--threshold) can be used to accelerate the computation by ignoring matches below the threshold. This is an approximation, because the output may miss results which are too close together (up to ~1000 datapoints apart). The results will be provided in the output file where each row is a tuple of (subsequence index, match index, correlation/distance)CLI flag:
--profile_type=ALL_NEIGHBORSpyscamp functions: selfjoin_knn, abjoin_knn
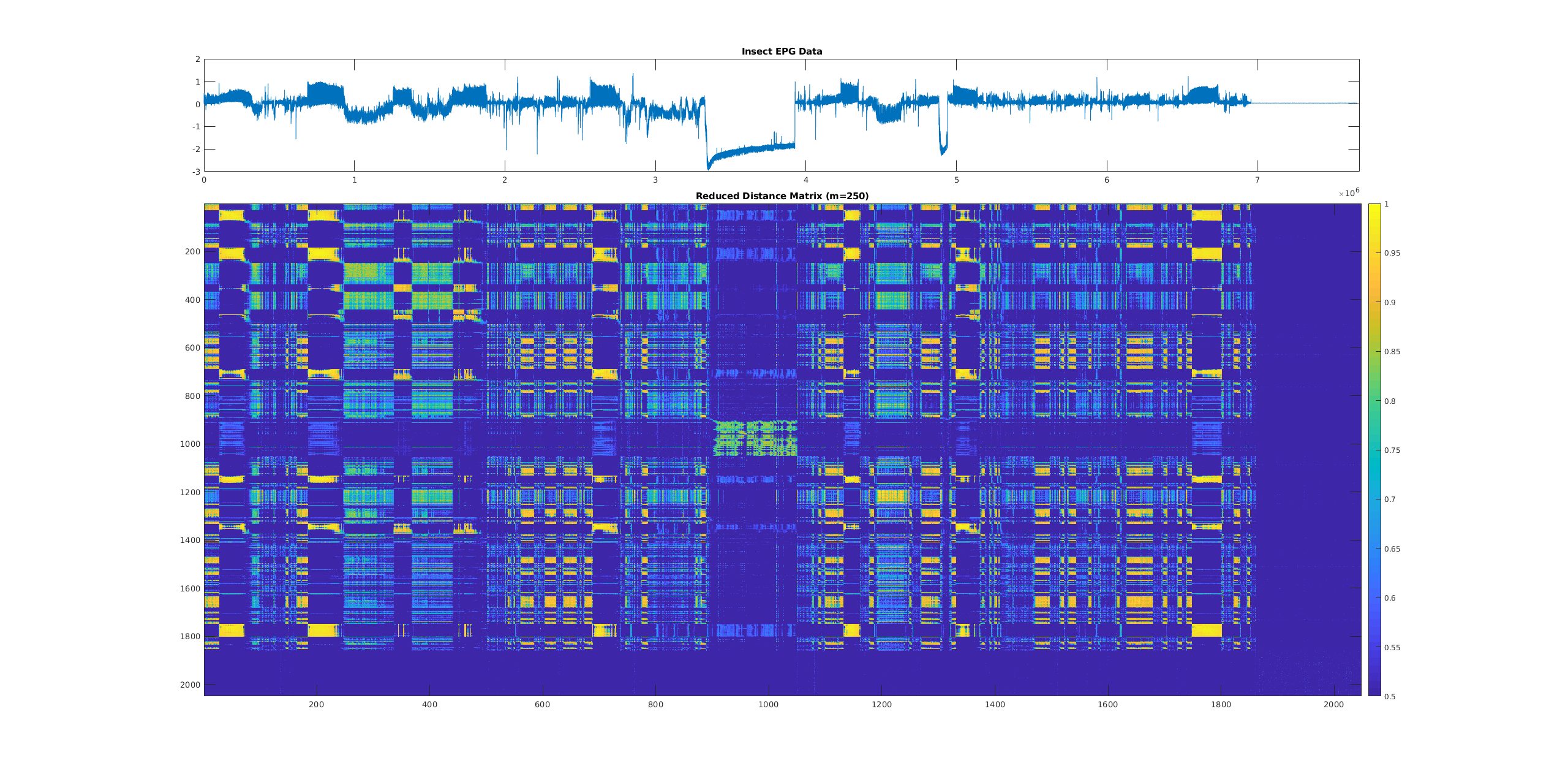
- Pooled distance matrix summary:
[EXPERIMENTAL, DISTRIBUTED UNSUPPORTED] This returns a max-pooled summary (see example below) of the distance matrix using the specified summary matrix height (
--reduced_height) and width (--reduced_width). When using GPUs there are limits to the resolution of the output. The output matrix height and width must be approximately 1000x smaller than the input size, otherwise you can get patchy results. If you have a small time series (~100K elements or less), it is recommended you use the CPU version for now as that is fast enough). Additionally, the entire output matrix must be small enough to fit in your system/GPU’s memory.CLI flag:
--profile_type=MATRIX_SUMMARYpyscamp functions: selfjoin_matrix, abjoin_matrix
Below is an example distance matrix summary, as you can see there is structure exposed via the visualization of the distance matrix.